Epigenetic clocks based on DNA methylation data are a type of biomarker that is useful for estimating biological age in population-based cohorts. However, usability has been limited by partially unreliable estimates due to this. noise in data. In a new study, we present a solution by introducing a method that reduces the noise in the epigenetic clocks for improved precision in longitudinal analyzes and clinical studies.
Biomarkers for aging can be obtained from cellular, molecular, functional and physiological measurements and used to study biological aging in humans. Many studies use the so-called "epigenetic clocks" based on DNA methylation data to analyze the relationship between biological aging and morbidity and mortality. Although these watches are currently considered to be the best predictors of biological age, they still include technical noise, leading to great variation in the measurements.
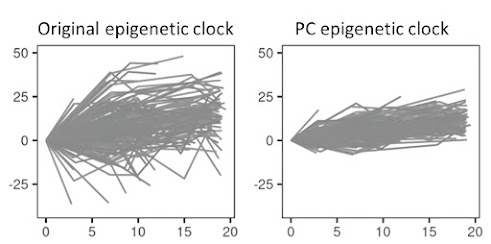
In order to make the clocks more useful for longitudinal analyzes and clinical studies, improved precision in the measurements is important. Here we present a method where principal component analysis (PCA) is used to separate noise from age-related signals. In this way, only biologically relevant signals are prioritized and the reliability of the PC watches is much higher compared to the original watches.















.jpg)