 |
| An artist's impression of E. coli, which infects over 150 million people worldwide. |
The University of Queensland-led study found a particular form of the bacteria – E. coli ST131 – had a previously unnoticed gene that made it highly resistant to commonly prescribed antibiotics.
Professor Mark Schembri, from UQ’s School of Chemistry and Molecular Biosciences, said this ‘resistance gene’ can spread incredibly quickly.
“Unlike gene transfer in humans, where sex is required to transfer genes, bacteria have genetic structures in their cells – called plasmids – that are traded quickly and easily between each other,” Professor Schembri said.
“This resistance gene is in one such plasmid and is swiftly making E. coli ST131 extremely resistant to widely prescribed fluoroquinolone antibiotics.
“These antibiotics are used to treat a wide range of infections, including urinary tract infections (UTIs), bloodstream infections and pneumonia.
“Importantly, this gene works with other resistance genes to achieve resistance at a level greater than the highest antibiotic concentrations that we can achieve during treatment.
“So we’re going to have to rethink our treatment plan, and strive to create antibiotics that can tackle these infections in spite of this antibiotic resistance mechanism.”
The findings have given the team the first clues to explain how antibiotic-resistant E. coli ST131 has emerged and spread so quickly around the world.
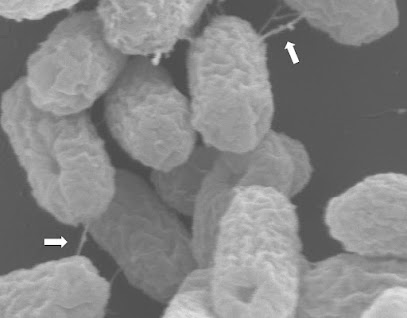 |
| E. coli ST131 superbugs captured transferring their antibiotic resistance plasmid through a syringe-like structure (indicated by arrows) |
E. coli causes more than 150 million infections each year, primarily urinary tract infections (UTIs).
It’s also one of the most common causes of sepsis, a disease that kills around 11 million people every year.
Now researchers’ sights are set on creating better treatments to stop E. coli ST131 infections in their tracks.
“We’ve lost a critical part of our armory to treat UTI and sepsis, but there’s still hope,” Professor Schembri said.
“Now that we understand the impact of this plasmid-mediated antibiotic resistance gene, we can devise more tailored treatment strategies.
“These might include new combinations of antibiotics, or even alternative non-antibiotic drugs that block E. coli ST131 infection.”
Study lead author, Dr Minh-Duy Phan, said this information could also be used to more efficiently track emerging resistance against critical last-line antibiotics.
“Resistance against antibiotics like carbapenems and polymyxins is emerging rapidly in some parts of the world, and we found the fluoroquinolone resistance gene we characterized in our study is often linked to such resistance,” Dr Phan said.
“Evolution has provided E. coli with this gene, but I’m confident that human ingenuity can still prevail against this deadly bacterium.”
The research has been published in Antimicrobial Agents and Chemotherapy
Source/Credit: University of Queensland
bio011322_04







.jpg)