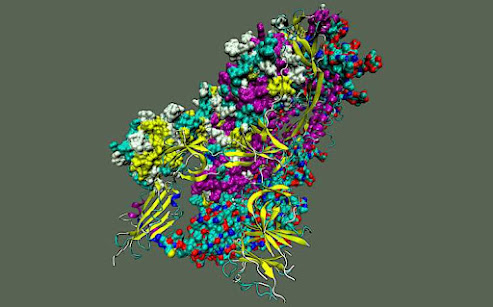
To explore the inner workings of severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2, researchers from the Department of Energy’s Oak Ridge National Laboratory developed a novel technique.
The team — including computational scientists Debsindhu Bhowmik, Serena Chen and John Gounley — ran molecular dynamics simulations of the novel virus that caused the COVID-19 disease pandemic on ORNL’s Summit supercomputer, an IBM AC922 system. The researchers then analyzed the output with a customized deep learning approach to produce a complete molecular picture of the “spike” protein on the virus’s surface.
This method enabled them to pinpoint specific flexible regions, which they studied in extreme detail to reveal promising therapeutic targets. Aiming for these targets could create more reliable treatment avenues that interrupt key structural transitions in the virus’s lifecycle while also supporting the body’s natural immune response.
“A better understanding of the spike protein could complement current COVID-19 vaccines by informing new treatments and providing insights into potential drug design,” Bhowmik said.













.jpg)
